Integration Site Analysis
Characterisation of genetic modifications for clone selection, genetic stability and lot release

What is INTEGRATION SITE ANALYSIS and why is it important?
As part of the development of a cell line for manufacturing, candidate clones will be selected based on a number of attributes including production yield, number of transgene copies, as well as the structure and location of the transgene in the host cell genome. Indeed, a certain level of genetic characterization of the cell line is expected by the regulatory authorities (as outlined in Cell Line Characterization).
However, developers can benefit from a deeper understanding of the integration site of the transgene when making clone selection. For example, transgenes located in areas of instability or regions that are prone to epigenetic silencing may lead to a significant reduction in manufacturing yields over the cell line’s manufacturing lifetime.
Classical integration site analysis methods such as fluorescence insitu hybridization (FISH) or southern blotting can give information on the integration location in the genome, but is still very low resolution. Sequencing the integration site provides single nucleotide resolution, however can be challenging, especially if the transgene is randomly integrated into the host genome. Methods such as long range PCR combined with Sanger sequencing are also employed, but are cumbersome and also require a priori knowledge of the integration site. More recently, NGS whole genome sequencing (WGS) has been used to characterize the integration site. However, WGS can be complex for cells with unstable genomes such as CHO. In addition, WGS tends to provide a lower level of sequence coverage, making conclusive identification of the integration site and its epigenetic status difficult.
A targeted short-read NGS method utilizes whole genome crosslinking to generate circular DNA fragments up to 100kb either side of a known primer site from the gene of interest. This method does not require any a priori sequence information of the insertion site, and good information is obtained on the junction site. However, due to the nature of short read sequencing this approach can struggle to identify concatemers, truncations and inverted repeats.
What we do
Modalities Tested:
- Cell banks for mAbs and recombinant biologics, Viral vectors, Cell therapies, Vaccines
- Cell therapy lots
- Gene therapy
- Cultured meat banks
Benefits of Long-Read iDTECT® NGS assay
✓ Validated method✓ Single nucleotide precision✓ Resolve concatemers, truncations and inverted repeats more easily than short-read NGS
Our Assay:
- iDTECT® Integration Site Analysis
INTEGRATION SITE ANALYSIS METHODS
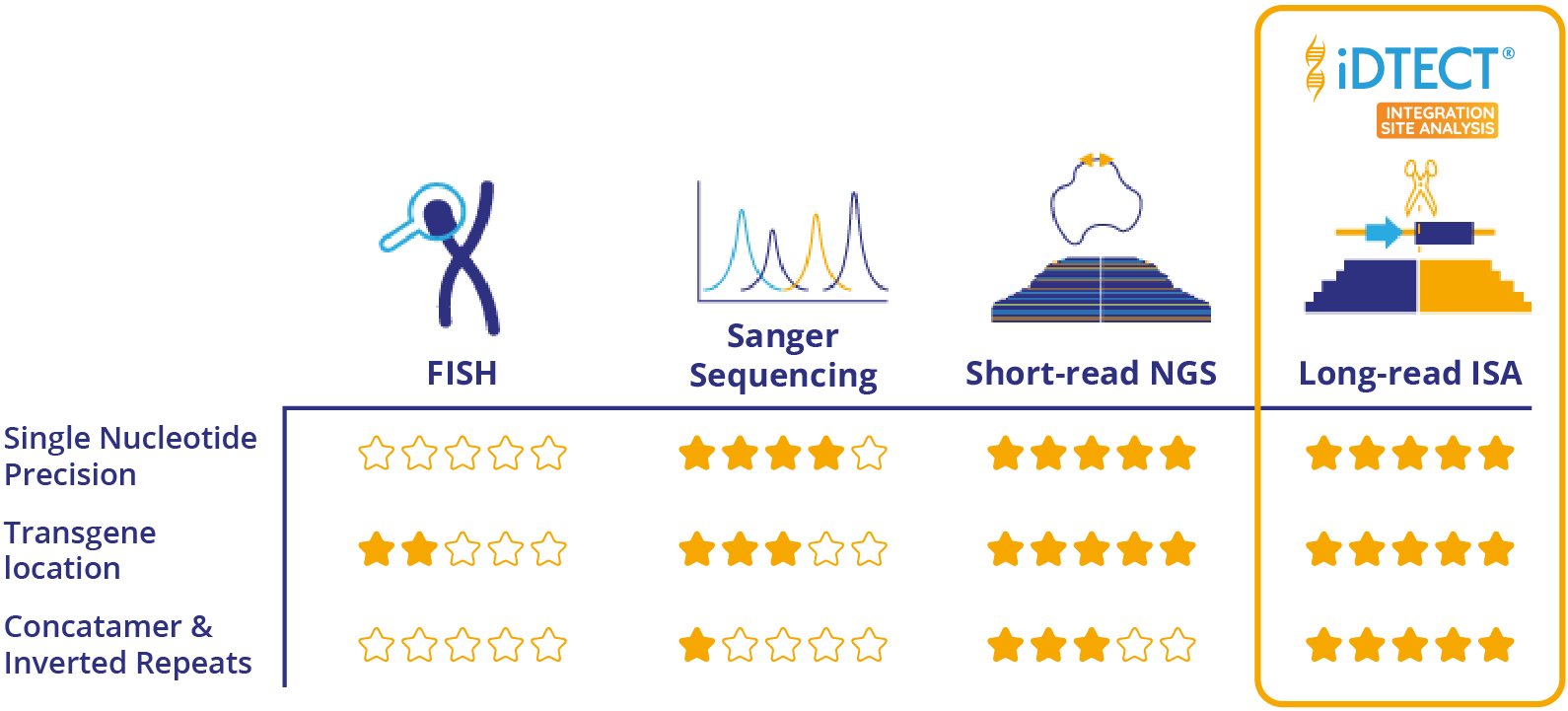
WHY THIS APPROACH
ICH: Scientific guideline Q5B analysis of the expression construct in cell lines
FDA: Characterization and Qualification of Cell Substrates and Other Biological Materials Used in the Production of Viral Vaccines for Infectious Disease Indications
Publication: Nanopore Cas9‐targeted sequencing enables accurate and simultaneous identification of transgene integration sites, their structure and epigenetic status.
PathoQuest Webinar
SAMPLE REQUIREMENTS
The specific sample requirements can depend on the source material and the desired outputs. We would therefore recommend having a consultation with one of our experts to discuss your exact requirements.
In general our sample requirements are:
- 5×10⁶ cells recommended (≥10µg DNA per sample)
- Storage and shipment at -80°C or on dry ice
- Cell pellets or cryo-preservation buffer
- Back up sample is recommended
Challenges solved
- Understanding complex transgene arrangements
- Low sample volumes
- Tight clinical timelines
- Reducing risk in cell banking with more information on integration site
OTHER SERVICES
Adventitious Virus Testing
Detection of viral contamination within the manufacturing process and beyond.
READ MORE
In Vivo Replacement
NGS as an ethical alternative to animals in biosafety testing and characterization.
READ MORE
Identity Confirmation
Genetic characterization of viral and plasmid products for release.
READ MORE
Cell Line Characterization
Biosafety screening and stability testing of manufacturing cells.
READ MORE
HLA Genotyping
Characterizing and screening for novel and emerging cell therapies.
READ MORE
Raw Material Testing
Screening of high-risk inputs such as animal products and media.
READ MORE
Contact us
U.S.
466 Devon Park Dr
Wayne, PA 19087
United States
E: contact@pathoquest.com
Sign up for our latest news
France
+33 (0)1 70 82 17 90
Biopark -Bâtiment B,
11, rue Watt
75013 Paris, France
How can PathoQuest help?
U.S.
466 Devon Park Dr
Wayne, PA 19087
United States
France
+33 (0)1 70 82 17 90
Biopark -Bâtiment B,
11, rue Watt
75013 Paris, France
E: contact@pathoquest.com
How can PathoQuest help?
Sign up for our latest news